This is the material for a poster presented at the BCA meeting in Cambridge, England, April 1-4, 1996.
1Bernstein + Sons, 5 Brewster Lane, Bellport, NY, USA,
2Protein Data Bank, Chemistry Dept., Brookhaven National
Laboratory, Upton, NY, USA,
3 San Diego Supercomputer Center, PO Box 85608, San Diego,
CA, USA
Work supported in part by US NSF, PHS, NIH, NCRR, NIGMS, NLM and DOE
under contract
DE-AC02-76CH00016 (for FCB) and US NSF grant no. BIR 9310154 (for
PEB).
The relationship between the new mmCIF format for macromolecular
structures and the
existing Protein Data Bank entry format is discussed, giving examples
of how to translate
between them.
A simple tabular concordance can be used for some tokens, and portions
of such a concordance
are presented. Much of the work of translation from PDB format to mmCIF
format has
been automated, though careful checking of the results is required and
considerable manual revision is necessary for some entries. Work on
automated translation from
mmCIF format to PDB format is in progress. The status of the programs
pdb2cif[BBB95]
and cif2pdb[BB96] is presented.
The Protein Data Bank format [PDB77, PDB95, PDB96] has been used for over 20 years
to archive
macromolecular data, is produced by many refinement programs, and is
used as an input
format by many applications. The pending adoption of the mmCIF
dictionary[FBB96] by the
IUCr, in response to the need to explicitly represent a larger amount
of data which can
be parsed by computer (necessary as the number of structures continues
to grow exponentially),
makes translation between mmCIF format and PDB format a pressing issue.
The two formats are different both in presentation and in content. The
PDB format
consists mainly of fixed format fields in an ordered set of records.
The new mmCIF format is one of a family of STAR (Self-Defining Text Archive
and Retrieval File [HS94]) formats
which uses a tag-value style of presentation and has very little
sensitivity to
the ordering of the information. The content of PDB entries is
organized around the presentation
of sets of atomic coordinates associated with chains and HET groups.
The content
of mmCIF data sets is organized around "entities" (discrete
chemical components).
With care, all the information of interest about a macromolecule can be
presented in either
format clearly and efficiently, but challenging problems arise in
moving between
the two formats. For example, given a PDB entry, identification of the
distinct entities needed for mmCIF may require looking for homologies
among the sequences of the chains.
As a further example, the PDB format treats a bifurcated sheet as two
distinct sheets
which happen to have certain strands in common, while mmCIF allows all
the strands involved to be represented as a single sheet. This requires
strand matching and alignment
to go from PDB format to mmCIF. Working in the other direction, from
mmCIF to PDB
format, the fixed fields of the PDB format limit the range of values
they may hold.
mmCIF [FBB96] uses a system of tags and values to describe a structure, as
shown in extract
giving the chain sequences from the pdb2cif conversion of PDB entry
4INS [DHH89]:
loop_
_entity_poly_seq.entity_id
_entity_poly_seq.num
_entity_poly_seq.mon_id
1 1 GLY 1 2 ILE 1 3 VAL 1 4 GLU 1 5 GLN
1 6 CYS 1 7 CYS 1 8 THR 1 9 SER 1 10 ILE
1 11 CYS 1 12 SER 1 13 LEU 1 14 TYR 1 15 GLN
1 16 LEU 1 17 GLU 1 18 ASN 1 19 TYR 1 20 CYS
1 21 ASN
2 22 PHE 2 23 VAL 2 24 ASN 2 25 GLN 2 26 HIS
2 27 LEU 2 28 CYS 2 29 GLY 2 30 SER 2 31 HIS
2 32 LEU 2 33 VAL 2 34 GLU 2 35 ALA 2 36 LEU
2 37 TYR 2 38 LEU 2 39 VAL 2 40 CYS 2 41 GLY
2 42 GLU 2 43 ARG 2 44 GLY 2 45 PHE 2 46 PHE
2 47 TYR 2 48 THR 2 49 PRO 2 50 LYS 2 51 ALA
Because tags are always given, the same information can be presented in
different
orderings. The Protein Data Bank [PDB96] uses a format with fixed fields
and is order-dependent.
Here is the sequence information from the PDB entry:
SEQRES 1 A 21 GLY ILE VAL GLU GLN CYS CYS THR SER ILE CYS SER LEU 4INS 170 SEQRES 2 A 21 TYR GLN LEU GLU ASN TYR CYS ASN 4INS 171 SEQRES 1 B 30 PHE VAL ASN GLN HIS LEU CYS GLY SER HIS LEU VAL GLU 4INS 172 SEQRES 2 B 30 ALA LEU TYR LEU VAL CYS GLY GLU ARG GLY PHE PHE TYR 4INS 173 SEQRES 3 B 30 THR PRO LYS ALA 4INS 174 SEQRES 1 C 21 GLY ILE VAL GLU GLN CYS CYS THR SER ILE CYS SER LEU 4INS 175 SEQRES 2 C 21 TYR GLN LEU GLU ASN TYR CYS ASN 4INS 176 SEQRES 1 D 30 PHE VAL ASN GLN HIS LEU CYS GLY SER HIS LEU VAL GLU 4INS 177 SEQRES 2 D 30 ALA LEU TYR LEU VAL CYS GLY GLU ARG GLY PHE PHE TYR 4INS 178 SEQRES 3 D 30 THR PRO LYS ALA 4INS 179
The major differences in syntax are as follows:
PDB and mmCIf formats agree simply and directly for some data items, such as cell parameters, and admit a simple tabular mapping, as shown by this extract from the concordance [B96] which is available as part of pdb2cif [BBB95]:
PDB Field Content Type of Transformation
and Related mmCIF field
CRYST1[1-6] CRYST1 NA
CRYST1[7-15] a equivalent to _cell.length_a
CRYST1[16-24] b equivalent to _cell.length_b
CRYST1[25-33] c equivalent to _cell.length_c
CRYST1[34-40] alpha equivalent to _cell.angle_alpha
CRYST1[41-47] beta equivalent to _cell.angle_beta
CRYST1[48-54] gamma equivalent to _cell.angle_gamma
CRYST1[56-66] sGroup equivalent to
_symmetry.space_group_name_H-M
CRYST1[67-70] z equivalent to _cell.Z_PDB
while other important macromolecular data descriptors, such as for sheets,
require complex transformations.
For more examples of mmCIF data and comparisons to PDB format, see the Macromolecular Crystallographic Information (mmCIF) Tutorial [PEB95].
pdb2cif [BBB95] is a program which converts PDB entries into mmCIF datasets. Most, but not all, common PDB record types are converted. The program cannot resolve some of the ambiguitites involved in the conversion.
The program produces summary warnings as comments at the end of each output CIF.
Unconverted records are captured in the AUDIT category warnings and converted records
should be examined carefully, especially for the following record types
COMPND, SOURCE, TITLE and CAVEAT
are merged into _struct.title without further parsing. A great deal of information
could be derived from the entries which use the PDB 1995 format description when
sufficient information for mapping of MOL_ID to entities is available.
END
records are ignored.
ENDMDL
records are ignored.
MODEL
is not supported. The suggested approach has been to use a separate data block for each
structure with a shared global header. This is not currently implemented.
REMARK
records currently are mapped without parsing. There is a great deal of information
in these records which can be parsed in more recent entries.
SIGATM and SIGUIJ records are ignored at this time. This will need to be addressed.
Additional data items for categories like _struct_topol will need to be added as they
evolve.
cif2pdb [BB96] is a program which converts mmCIF datasets into "pseudo-PDB" format (a format sufficiently similar to standard PDB format to be accepted by most applications). It is written in Fortran using CIFtbx2 [HB96]. In its present form, it is able to produce HEADER, SCALE, ORIGX, CRYST1, and ATOM/HETATM/TER, which provides sufficient information to drive RASMOL [S94] to produce drawings from mmCIF datasets. The images shown here were produced from CIF's converted to PDB format by cif2pdb and then rendered by RASMOL. Here is are images of 4INS [DDH89], 1ACE [SHS91] and DDF040 [GWB95] :
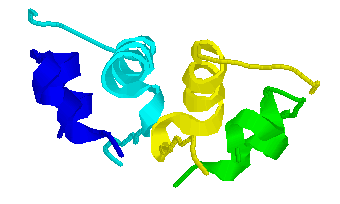
4INS is a PDB entry for Insulin. This image was created by processing 4INS through pdb2cif to produce an mmCIF data set, then through cif2pdb to create a pseudo-PDB entry, and finally through RASMOL.
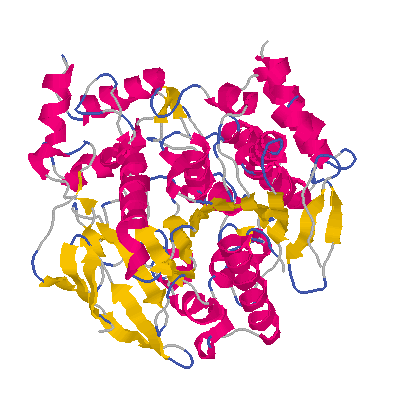
1ACE is a PDB entry for Acetylcholinesterase. This image was created by processing 1ACE through pdb2cif to produce an mmCIF data set, then through cif2pdb to create a pseudo-PDB entry, and finally through RASMOL.
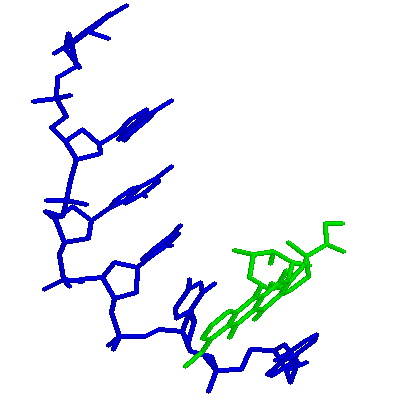
DDF040 is an example mmCIF data set for a DNA-Drug complex structure. It was "born" as an mmCIF data set. This is image was created by processing DDF040 through cif2pdb to create a pseudo-PDB entry, and finally through RASMOL.
There are many useful sites on the World Wide Web where information, tools and software related to the Protein Data Bank and mmCIF can be found. The following are good starting points for exploration:
The Protein Data Bank provides access to entries, software and documenation with a browser and an on-line PDB format description at:
with a mirror for European users at:
The Nucleic Acid Database Project provides access to its entries, software and documentation, with an mmCIF page giving access to the dictionary and mmCIF software tools at:
with a mirror for European users soon to be established at:
Information and Software for STAR and CIF can be found at:
and at
Tutorials on mmCIF and the relationship to PDB format can be found at: