
![[IUCr Home Page]](ciftbx/.logos/iucrhome.jpg)
![[CIF Home Page]](ciftbx/.logos/cifhome.jpg)
![[ciftbx]](ciftbx/.logos/ciftbxButton.jpg)
![[cif2pdb]](cif2pdb/.logos/cif2pdbButton.jpg)
![[pdb2cif]](pdb2cif/.logos/pdb2cifButton.jpg)



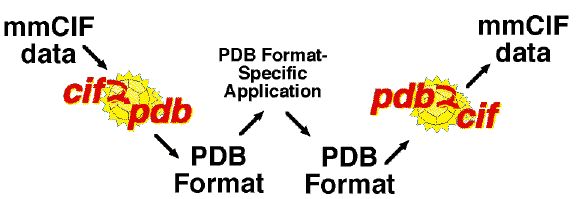
There are many approaches to code conversion to allow applications to use CIF. An application which reads other data formats may be "converted" immediately by use of an appropriate preprocessing filter to convert the data from CIF to a data format already known to the application. An application which writes other data formats may similarly be "converted" by use of an appropriate post-processing filter. For example, if the application is question is PDB format specific, then it may be possible to adapt it for use with mmCIF by using cif2pdb [BB96] as a preprocessing filter and pdb2cif [BBB98] as a post-processing filter.
Depending on the application and the nature of the environment, there are different filtering programs to consider. The NDB's MAXIT handles a wide range of conversions in a production environment involving a variety of formats. The program QUASAR [HS93] provides users of core CIF data with a program which can extract tags and values from a CIF with the tags in a specified ordering. This can unburden an application from the chores of accepting data the application considers extraneous and from having to deal with arbitrary orderings of columns of data. The program cif2cif (see the CIFtbx release) can perform similar tasks for mmCIF as well as core CIF data. The approach of using filters has much to recommend it, especially when conversion of an application must be performed quickly.
However, there are cases where internal conversion of an application is preferable. Because the data involved need not be reprocessed, converted code is often faster than equivalent code working with filter programs. The code within an application can be tuned to the internal data structures and coding conventions of the application.
The approach to internal conversion depends on the language, data structures and operating environment of the application. A few years ago, the precise details of language version and operating system would have been major stumbling blocks to conversion. Today, however, almost every platform supports a variation of the Unix application programming interface and many languages have viable interfaces to C and/or C++. Therefore it is often feasible to consider use of C, C++ or Objective-C libraries, even for Fortran applications. For non-Fortran libraries we refer you to the work of J. Westbrook and of Phil Bourne.
If an application in Fortran is to be converted with a Fortran-based library, the package CIFtbx [HB96] is one possible solution. Even if one chooses to produce a new interface library from scratch, say, for maximal efficiency or to ensure a full personal understanding of CIF, this package provides a viable starting point and examples to consider.
One of the major considerations in dealing with CIF from within Fortran is that a valid CIF may present data in any ordering. Fortran does not provide facile memory for the necessary explicit or implicit internal sorting. This makes it natural to transfer CIFs being read to a disk-based random access file. CIFtbx does this each time it opens a CIF. The user never works directly with the original CIF data set. This provides a clean and simple interface for reading, but slows all read access to CIFs. Compromises are often necessary, with critical tables handled in memory rather than on disk, but that may forces changes in dimensions and then recompilation when dictionaries or data sets become larger than anticipated.
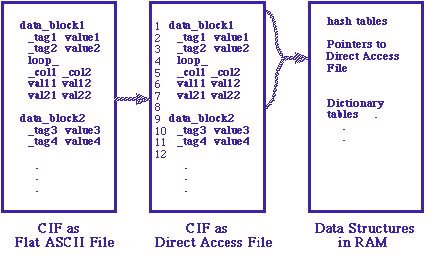
Writing a CIF is simpler than reading a CIF because the items in a CIF can be written in any convenient order. In most cases the data may be written in whatever order is natural for the application.
However, both while reading and while writing a CIF, it is desirable to check tags against the appropriate dictionaries. If the dictionaries are kept as disk files, this can slow execution, so the simplest practice is to extract the necessary information from the dictionaries and hold it in memory. CIFtbx performs this function for you.
Whether the choice is to use filters or to rewrite the application, CIF and mmCIF are well-supported by tools for the Fortran programmer.
For a discussion of the rationale behind CIFtbx, cif2cif and Cyclops, see Reading, Writing and Validating CIFS using CIFtbx2, cif2cif and Cyclops by S. R. Hall and H. J. Bernstein.
For a discussion of the rationale behind the program cif2pdb, see cif2pdb: Translating mmCIF Data into PDB Entries by F. C. Bernstein and H. J. Bernstein.
For a discussion of the rationale behind pdb2cif, see Translating PDB Entries into mmCIF by P. E. Bourne, F. C. Bernstein and H. J. Bernstein.
There are many useful sites on the World Wide Web where information, tools and software related to CIF, mmCIF and the PDB can be found. The following are good starting points for exploration:
The International Union of Crystallography (IUCr) provides access to software, dictionaries, policy statements and documentation relating to CIF and mmCIF at:
with mirror sites at:The Nucleic Acid Database Project provides access to its entries, software and documentation, with an mmCIF page giving access to the dictionary and mmCIF software tools at:
with mirror sites at:The Protein Data Bank provides access to entries, software and documentation with a browser, and an on-line PDB format description at:
with mirror sites at many locations (see http://www.pdb.bnl.gov/pdb-docs/mirror_sites.html).
Tutorials on mmCIF and the relationship to PDB format can be found at: http://www.sdsc.edu/pb/cif/tutorials.html
Here are direct links to copies of the IUCr CIF home page, the NDB's mmCIF home page, pdb2cif, cif2pdb and CIFtbx (with Cyclops and cif2cif).
|
United States
| |||||
| NDB, Rutgers, NJ | mmCIF | pdb2cif | cif2pdb |
CIFtbx... | |
| SDSC, San Diego, CA | CIF | mmCIF | pdb2cif | cif2pdb |
CIFtbx... |
|
United Kingdom
| |||||
| IUCr, Chester | CIF | pdb2cif | cif2pdb | CIFtbx... | |
| EBI, Hinxton | mmCIF | pdb2cif | cif2pdb |
CIFtbx... | |
|
France
| |||||
| U. P. et M. Curie, Paris | CIF | pdb2cif | cif2pdb | CIFtbx... | |
| Sweden | |||||
| U. of Stockholm | CIF | pdb2cif | cif2pdb | CIFtbx... | |
|
South Africa
| |||||
| U. of the Witwatersrand | CIF | pdb2cif | cif2pdb | CIFtbx... | |
|
Japan
| |||||
| NIBH, Ibaraki | mmCIF | pdb2cif | cif2pdb |
CIFtbx... | |
|
Australia
| |||||
| UWA, Nedlands | STAR/CIF | pdb2cif | cif2pdb |
CIFtbx... | |
Herbert J. Bernstein (yaya@bernstein-plus-sons.com)